Kuldeep Singh1, Satish Kumar2 and Pawan Kaur3*
1Department of Computer Science, FGM Govt. College, Aadampur-125052, Hisar, Haryana, India
2Department of Computer Applications, Panjab University SSG Regional Centre, Hoshiarpur-146001, Punjab, India
3Department of Biotechnology, Chaudhary Devi Lal University, Sirsa-125055, Haryana, India
Corresponding Author Email: pawankaurnano@gmail.com
DOI : http://dx.doi.org/10.13005/ojcst/09.03.08
Article Publishing History
Article Received on :
Article Accepted on :
Article Published : 27 Dec 2016
Article Metrics
ABSTRACT:
Powdery mildew disease of beans in India causes major economic losses in agriculture. For sustainable agriculture detection and identification of diseases in plants is very important. In this review, we are trying to identify the powdery mildew disease of beans crop by using some image processing and pattern recognition techniques and comparing with molecular and spectroscopic techniques. Early information on crop health and disease detection can facilitate the control of diseases through proper management strategies. The present review recognizes the need for developing a rapid, cost-effective, and reliable health monitoring techniques that would facilitate advancements in agriculture. These technologies include image processing and pattern recognition based plant disease detection methods
KEYWORDS:
beans; powdery mildew; detection; image processing; pattern recognition
Copy the following to cite this article:
Singh K, Kumar S, Kaur P. Detection of Powdery Mildew Disease of Beans in India : A Review. Orient.J. Comp. Sci. and Technol;9(3)
|
Copy the following to cite this URL:
Singh K, Kumar S, Kaur P. Detection of Powdery Mildew Disease of Beans in India : A Review. Orient. J. Comp. Sci. and Technol;9(3). Available from: http://www.computerscijournal.org/?p=4051
|
Introduction
Pulse production has been stagnant in Asian countries leading to steady decline in per capita availability over the past 50 years. In India 13.00 – 15.00 million metric tons (mmt) of peas and pulses produced which is about 25% of the global production. India is the largest producer as well as major importer of peas/pulses in the World. Pulse is a common name for all varieties of dry peas, lentils, beans and chickpeas. Almost all varieties of pulses, except lupins, faba beans and dun peas, are grown in India.
Beans are intricately woven into the fabric of human history. The primary ‘everlasting cultures’ advanced when hunter-gatherers and nomadic persons started tilling the earth, establishing programs of agriculture and beans were among the first cultivated crop. Among beans soybean (Glycine max (L.) Merr.) and peanut (Arachis hypogea L.) are the 3rd most essential food legume crop worldwide [1]. Cultivated beans are consumed in form of dry edible beans (French bean (Rajma), soybean, mung bean, lima bean and azuki bean) as the mature dry seed after rehydration, and snap beans (e.g., green, string, Haricot bean) as fleshy immature pods. Dry beans are further divided into distinct market classes based on seed characteristics and snap bean classes are based on pod characteristics and plant type [2].
Powdery Mildew Diseases Of Beans
Diseases are often encountered through the growing stage, the production stage which is most favorable stage for various plant pathogens like crop residue-borne fungi and other soil-surface-inhabiting microbes and postharvest storage [3, 4]. The most vital factor for the development of certain plant pathogens and plant diseases is that the presence of wetness (moisture) at the soil-air interface or within the top few millimeters of soil. [5, 6]. Soil surface moisture, usually related to the precipitation events or dew formation in the canopy [7, 8], is plausible to be one of the most important environmental factor affecting the growth of plant pathogens to cause many bacterial and fungal plants diseases [2].
Powdery mildew, a fungal disease of beans, is caused by many different species of fungi in the order Erysiphales and affects a wide range of beans plants. It is a foliar disease of beans, as its symptoms are quite distinctive in form of white powdery spots on the leaves and stems of the infected plants (Figure1). The most affected part in the infected plants is the lower surface of leaves, but the mildew can appear on any above-ground part of the plant. Disease progresses in the form of white spots of asexual spores that get larger and denser and may spread up and down the stem of the plant [13].
Here, in this review, we are focusing on powdery mildew disease i.e. foliar diseases of beans and most critical leaf surface conditions; favorable temperature (22-27 °C) and leaf surface humidity (High relative humidity at night, Low relative humidity during day) are required for occurrence of powdery mildew disease [9]. These favorable conditions cause greater the risk of infection and the greater the number of infections per leaf. Even powdery mildew diseases have been reported to be more severe with prolonged periods of these critical conditions [10-12]. Thus, disease management prediction requires leaf wetness assessment.
Management of Powdery Mildew Disease
In agriculture, there are various methods that can be applied for controlling powdery mildew disease i.e. fertilizers, chemical methods, genetic resistance, and careful farming methods. It is important to be aware of powdery mildew and its management as the resulting disease can significantly reduce crop yields [14].
Carefully timing application and rate of nitrogen as fertilizer (less than 70 pounds/acre) can be used to eliminate the progress of disease by altering planting density. Another ways to manage disease are crop rotation, eliminating the presence of volunteer in agriculture field, use of highly resistance varieties [15]
Chemical control is possible with fungicides such as triadimefon and propiconazole. Bennet et al. (1984) used calcium silicate slag for treatment of powdery mildew disease of wheat that helps the plant cells to defend against fungal attack by degrading haustoria and by producing callose and papilla [16]. Some studies have shown milk’s effectiveness as comparable to some conventional fungicides [17], and as better as benomyl and fenarimol at higher concentrations [18]. Some home gardeners and small scale farmers have been used milk for the treatment of mildew disease, by diluting with water and sprayed on infected plants at the initial stage, or as a preventative measure, with repeated weekly application on the plants.
Some agricultural universities and institutes have been releasing resistance varieties of many crops that can be an economically and efficient method in disease reduction. The most common and serious disease-resistant varieties should be selected on the basis of local adaptability and high yield potential [19].
Plant Disease Detection
Molecular and Spectroscopic Techniques
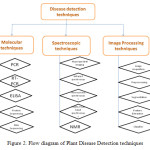
Indian farming is significantly affected by plant diseases that cause major production and economic losses as well as negative agronomic impact on Indian agriculture [20]. Plants infected with disease or not, are confirmed by using disease detection molecular techniques. Molecular techniques available for plant disease detection are polymerase chain reaction (PCR), enzyme-linked immunosorbent assay (ELISA), protein sequencing and nucleic acid (DNA) sequencing etc. [21-26] as shown in figure 2.
Plants diseases have been detected by applying unique disease monitoring methods, which are known as the spectroscopic and imaging techniques. Development of such technologies to implement an experimental tool for a large scale disease monitoring in the field, has been involved in current research activities. Symptomatic or asymptomatic plants diseases have been detected by various spectroscopic and imaging techniques. Some methods: fluorescence imaging [27-29], multispectral or hyperspectral imaging [30-32], infrared spectroscopy [33, 34], fluorescence spectroscopy [35-37], visible/multiband spectroscopy [38-40], and nuclear magnetic resonance (NMR) spectroscopy [41] have been used for diseases detection as shown in figure 2.
Powdery mildew (Erysiphe graminis sp. tritici) in wheat (Triticum aestivum L.) was identified by using leaf reflectance measurements [42]. Similarly, Lorenzen and Jensen [43] used spectral reflectance measurements to make an early diagnosis of symptoms on barley leaves infected by cereal powdery mildew. However, the reduction in chlorophyll content is not the common point in disease related stress
Sasaki et al. (1998) have distinguished diseased cucumber leaves from healthy leaves at an early infection stage, based upon the spectral reflectance of the leaves in the 500, 600 and 650 nm wavebands. The classification performance was 90% [44].
In spite of availability of these techniques, there is a demand for a fast, sensitive, and selective method for the rapid detection of plant diseases. In the present review paper, molecular methods and spectroscopic methods of plant disease detection are not discussed in detail as a number of review papers are available in the literature, we are focusing on image processing techniques and describing more in details.
Image Processing and Pattern Recognition techniques
Image processing technology plays important role in the agricultural research in detection of plant diseases. Computer image processing technology was developed in the 1960 of the 20th century, but applied in the 1970s in the area of the production and processing of agricultural products [20].
In plant sciences, thousands of images are required for experimental analysis like area of lesions on the leaves, area damaged by insects, scoring quantitative and qualitative traits, etc. All these experiments are analysed manually or with individual software packages. It is very time consuming, tremendous amount of works, problem with analytical accuracy of results and subjectiveness rising from different individuals. So to overcome these issues, plant biologist need efficient software like image processing that can automatically extract and analyze significant content.
Some scientists developed new methods based on image processing and pattern recognition techniques for fast and accurate grading of plant diseases as shown in figure 2. Many research works have been published regarding the advancements of image processing for feature extraction, segmentation, preprocessing and classification.
A novel approach is proposed [45] for integrating image analysis technique into diagnostic expert system. A CLASE (Central Lab. of Agricultural Expert System) diagnostic model is used to manage cucumber crop. This expert system diagnoses different disorders such as Leaf miner, Powdery and Downey disease by analyzing four image processing phases: enhancement, segmentation, feature extraction and classification. This proposed methodology reduced error and enhance experimental accuracy.
Recent developments in agricultural science have lead to a demand for a new era of automated methods of plant disease detection. It is necessary that the plant disease detection techniques should be rapid, specific, and sensitive for detection at the early stage of the disease [20].
An automated system has been developed using algorithm such as bounding box method chain code technique and moment analysis to recognize and classify fungi disease [46]. Severity of Rust disease on Soybean, also measured by Sobel operator to find out spot edge and plant disease severity by calculating the quotient of disease spot area and leaf area [47]. Kaundal et al. (2006) proposed a prediction approach based on support vector machines for developing weather based prediction models of plant diseases [48]. They compared the performance of various techniques like conventional multiple regression, support vector machine and artificial neural network (back propagation neural network, generalized regression neural network). It was observed that Support Vector Machine based regression approach is useful for disease management which could be a better description of the relationship between the environmental conditions and disease level.
Back propagation neural network also proposed for recognition of leaves specificity [49]. It was proved that species of a plant can be specified by back propagation neural network using just shape of leaf image. Thinning algorithm and prewitt edge detection were used to find leaf tokens as input to back propagation algorithm. It was concluded that an expert system can be developed to recognize various plant disease related to leaves like recognition of damaged leaves due to insects and fungal or bacterial diseases of leaves. Neural network approach for segmentation of agricultural landed fields in remote sensing data was proposed [50] in which neural network algorithm based on back propagation is used for segmentation of the color images of crop field infected by diseases that changes usual color of plants.
Pixia et al [51] used image processing and recognition technologies for detection of the disease of cucumber downy mildew, powdery mildew and anthracnose. In this study segmentation of leaf spot disease, feature extraction of lesion site, median filtering method of filtering noise were used to detect diseases of images and accuracy rates more than 96% was observed based on the shortest distance methods.
Digital image processing and pattern recognition techniques used for detection powdery mildew disease in the betelvine plants [52]. Data of digital images of the leaves were collected from betelvine plants at various stages using a high resolution digital camera. Analysis of images of the leaves was done using the image processing toolbox in MATLAB. For providing standard patterns of the digital images, preprocessed images were separated using RGB encoding technique the red, green and blue components. These patterns and images of various healthy betelvine leaves at different stages were collected periodically and stored in the system for analysis. Based on these patterns, it was identified whether test leaves were affected by powdery mildew disease or not. Finally using pattern recognition techniques, the powdery mildew disease can be identified at early stage before it spreads to entire crop.
A method was presented to monitor plant disease which caused by spores [53]. For analysis and processing, the color image was first converted in to gray image (the gray-level correction, histogram generation, feature extraction, image sharpening etc.). Then preprocessing of grey image was done by edge enhancement using the Median filter and canny edge algorithm in order to remove low frequency components. After thresholding binary image obtained was processed by using morphological features like dilation, erosion, opening etc. It was found that using this method, microscopic plant diseases, optics stripe counting and the chromosome counting etc. can be easily detected. Seiffert et al. (2005) also developed new pattern recognition software (HyphArea) for automated quantitative analysis of hyphal growth rates of powdery mildew fungi on plant. By using this software, the development of fungal pathogens can be quantified easily at the level of spore germination or penetration. However, the exact quantification of hyphal growth rates after initial, successful host invasion is much more difficult [54] but this pattern recognition tool can be helpful for disease detection at early stages.
After studying these methods and techniques, we can draw some conclusions that various molecular and hyper spectral remote sensing techniques are used to identify and detect plant diseases but there are more studies are needed to explore the spectral response characteristics of crops under disease stress of various levels in field and weather conditions. The image processing and pattern recognition techniques are used to detect disease of crops by feature extraction of infected images and healthy images to distinguish there features on the basis of shape, texture and colour.
On the basis of these techniques, powdery mildew disease can be identified by following features:
- At initial stage, white powdery fungus grows on the upper leaf surface of the lower leaves
- Next stage, fungus spread in the form of mats is called mycelium and appears as white or grayish cottony patches on leaves, buds and stems.
- Final stage, fruiting bodies (cleistothecia) appear as small black or brown specks on the mycelia mats.
Conclusion
In this review, there are so many methods used to identify, classify and detect plants disease and using image processing and pattern recognition techniques, preventive action can be taken well in advance such that the entire plantation can be protected before spreading of the disease. The method of detecting plant disease should be cost effective and non-destructive. Image processing techniques fulfil these characteristics as it not only requires the simple image data of the leaf samples, but also eliminates subjectivity of traditional methods and human induced errors. Early stage detection will helps to farmers to decide the specific quantity for fungicide application which reduces the cost and environmental pollution.
However, a higher number of experiments and improvements in the image-processing subroutine are needed to validate the method. In future we can utilize other principal component classifier such as neural network, back propagation and Support Vector Machine. These analysis methods and sensor specificities can be transferred and generalized for other plant-pathogen systems. Moreover, the technologies may be also used in plant pathology for investigating the effect of pathogenesis on the initial level.
References
- Singh, S. P. Production and utilization., “In: S. P. Singh (ed.). Common Bean Improvement in the Twenty-First Century”. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp, 1-24, 1999.
CrossRef
- Myers, J. R. and Baggett, J. R., “Improvement of snap beans. In: S. P. Singh (ed.) Common Bean Improvement in the Twenty-First Century”. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp, 289-330, 1999.
CrossRef
- Cook, R.J.; Duniway, J.M., “Water relations in the life-cycles of soilborne plant pathogens. In Water Potential Relations in Soil Microbiology”. Parr, J.F., Gardner, W.R., Elliott, L.F., Eds.; Soil Science Society of America: Madison, WI, 1981; Vol. 9, pp. 119-139.
- Rotem, J., “Climatic and weather influences on epidemics. In Plant Disease: An Advanced Treatise”. Horsfall, J.G., Cowling, E.B., Eds.; Academic Press: New York, 1978; Vol. 2, pp. 317-337.
- Swan, L.D.; Backhouse, D.; Burguess, L.W., “Surface soil moisture and stubble management practice effects on the progress of infection of wheat by Fusarium pseudograminearum”. Aust. J. Exp. Agr., vol.40, pp.693-698, 2000.
CrossRef
- Zhang, W., and Pfender, W.F., “Effect of residue management on wetness duration and ascocarp production by Pyrenophora triticirepentis in wheat residue,” Phytopathology,Vol. 82, pp. 1434-1439, 1992.
CrossRef
- Rosenberg, N.J.; Blad, B.L.; Verma, S.B., “Microclimate: The Biological Environment,” 2nd Ed.; John Wiley & Sons: New York, NY, U.S.A., 1983.
- Sirjusingh, C.; Sutton J.C., “Effects of wetness duration and temperature on infection of geranium by Botrytis cinerea”. Plant Dis. Vol. 80, 160-165. 1996,
CrossRef
- Agrios, G. N. Plant Pathology, 3rd Ed.; Academic Press, Inc.: New York, NY, U.S.A., 1988.
- Fidanza, M.A.; Dernoeden, P.H.; Grybauskas A.P. Development and field validation of a brown patch warning model for perennial ryegrass turf. Phytopathology, Vol. 86, pp. 385-390,1996.
CrossRef
- Giesler, L.K.; Yuen, G.Y.; Horst, G.L., “The microclimate in tall fescue turf as affected by canopy density and its influence on brown patch disease.” Plant Dis., Vol. 80, pp. 389-394,1996.
CrossRef
- Stavely j R, Freytag,G.F., Steadman J R, Schwartz H. F., The Bean rust workshop. Bean Improv. Coop. Ann. Rept. Vol. 26: iv-vi; 1983.
- Huang, X.Q., Hsam S. K. L., Zeller, F. J., Wenzel, G., Mohler, V., “Molecular mapping of the wheat powdery mildew resistance gene Pm24 and marker validation for molecular breeding.” Theoretical and Applied Genetics, Vol. 101. pp 407-414, Aug.2000.
CrossRef
- Maloy, Otis and Debra Inglis., “Powdery Mildew,” Washington University extension, Diseases of Washington Crops. 1993.
- Lipps, Patrick E. (n.d). “Powdery Mildew of Wheat,” The Ohio State University Extension. http://ohioline.osu.edu/ac-fact/0010.htmltm.
- Bennet, Fiona G. A., “Resistance to powdery mildew in wheat: a review of its use in agriculture and breeding programmes.” Plant Pathology, Vol. 33, pp. 279–300, Sept. 1984.
CrossRef
- DeBacco, Matthew. “Compost Tea and Milk to Suppress Powdery Mildew (Podosphaera xanthii) on Pumpkins and Evaluation of Horticultural Pots Made from Recyclable Fibers Under Field Conditions”. MSc Thesis,University of Connecticut, 16 May 2013.
- Bettiol, Wagner., “Effectiveness of cow’s milk against zucchini squash powdery mildew (Sphaerotheca fuliginea) in greenhouse conditions”. Crop Protection, Vol. 18, pp. 489–492, Sept.1999.
CrossRef
- Bélanger R. R, Nicole Benhamou, and Menzies, J. G., “Cytological Evidence of an Active Role of Silicon in Wheat Resistance to Powdery Mildew (Blumeria graminis f. sp. tritici).” Phytopathology, Vol. 93, pp. 402-407, April, 2003.
CrossRef
- López, M.M., Bertolini, E., Olmos, A., Caruso, P., Gorris, M.T., Llop, P., Penyalver, R., Cambra, M., “Innovative tools for detection of plant pathogenic viruses and bacteria”. International Microbiology, Vol. 6, pp. 233–243, 2003.
CrossRef
- Das, A.K., “Rapid detection of Candidatus Liberibacter asiaticus, the bacterium associated with citrus Huanglongbing (Greening) disease using PCR”. Current Science, Vol. 87, pp. 1183–1185, 2004.
- Li, W., Hartung, J.S., Levy, L., “Quantitative real-time PCR for detection and identification of Candidatus Liberibacter species associated with citrus Huanglongbing”. Journal of Microbiological Methods, Vol. 66,pp. 104–115,2006.
CrossRef
- Prithiviraj, B., Vikram, A., Kushalappa, A.C., Yaylayam, V., “Volatile metabolite profiling for the discrimination of onion bulbs infected by Erwinia carotovorassp. carotovora, Fusarium oxysporum and Botrytis allii”. European Journal of Plant Physiology. Vol.110, pp. 371–377, 2004.
- Ruiz-Ruiz, S., Ambrós, S., Carmen Vives, M., Navarro, L., Moreno, P., Guerri, J., “Detection and quantification of Citrus leaf blotch virus by TaqMan real-time RTPCR”. Journal of Virological Methods. Vol. 160, pp. 57–62, 2009.
CrossRef
- Saponari, M., Manjunath, K., Yokomi, R.K., “Quantitative detection of Citrus tristeza virus in citrus and aphids by real-time reverse transcription-PCR (TaqMan®).” Journal of Virological Methods. Vol. 147, pp. 43–53, 2008.
CrossRef
- Yvon, M., Thébaud, G., Alary, R., Labonne, G., “Specific detection and quantification of the phytopathogenic agent ‘Candidatus Phytoplasma prunorum”. Molecular and Cellular Probes. Vol. 23, pp. 227–234, 2009.
CrossRef
- Bravo, C., Moshou, D., Oberti, R., West, J., McCartney, A., Bodria, L., Ramon, H., “Foliar disease detection in the field using optical sensor fusion. Agricultural Engineering International: the CIGR Journal of Scientific Research and Development,” Manuscript FP 04 008, Vol. VI. 2004.
- Moshou, D., Bravo, C., West, J., Wahlen, S., McCartney, A., Ramon, H., “Automatic detection of ‘yellow rust’ in wheat using reflectance measurements and neural networks.” Computers and Electronics in Agriculture, Vol. 44, pp.173–188, 2004.
CrossRef
- Chaerle, L., Lenk, S., Hagenbeek, D., Buschmann, C., Van Der Straeten, D., “Multicolor fluorescence imaging for early detection of the hypersensitive reaction to tobacco mosaic virus”. Journal of Plant Physiology. Vol. 164, pp.253–262, 2007.
CrossRef
- Moshou, D., Bravo, C., Oberti, R., West, J., Bodria, L., McCartney, A., Ramon, H., 2005. “ Plant disease detection basedondata fusion of hyper-spectral and multi-spectral fluorescence imaging using Kohonen maps. Real-Time Imaging” Vol. 11, pp.75–83, 2005.
CrossRef
- Shafri, H.Z.M., Hamdan, N., “Hyperspectral imagery for mapping disease infection in oil palm plantation using vegetation indices and red edge techniques.” American Journal of Applied Sciences, Vol. 6, pp. 1031–1035, 2009.
CrossRef
- Qin, J., Burks, T.F., Kim, M.S., Chao, K., Ritenour, M.A., “ Citrus canker detection using hyperspectral reflectance imaging and PCA-based image classification method”. Sensing and Instrumentation for Food Quality and Safety. Vol.2, pp. 168–177, 2008.
CrossRef
- Spinelli, F., Noferini, M., Costa, G., “Near infrared spectroscopy (NIRs): Perspective of fire blight detection in asymptomatic plant material.” Proceeding of 10th International Workshop on Fire Blight. Acta Horticulturae, Vol.704, pp.87–90, 2006.
CrossRef
- Purcell, D.E., O’Shea, M.G., Johnson, R.A., Kokot, S., “Near-infrared spectroscopy for the prediction of disease rating for Fiji leaf gall in sugarcane clones”. Applied Spectroscopy. Vol. 63, pp. 450–457, 2009.
CrossRef
- Marcassa, L.G., Gasparoto, M.C.G., Belasque Junior, J., Lins, E.C., Dias Nunes, F., Bagnato, V.S., “Fluorescence spectroscopy applied to orange trees”. Laser Physics. Vol. 16, pp. 884–888, 2006.
CrossRef
- Belasque, L., Gasparoto, M.C.G., Marcassa, L.G., “Detection of mechanical and disease stresses in citrus plants by fluorescence spectroscopy.” Applied Optics, Vol. 47, pp. 1922–1926, 2008.
CrossRef
- Lins, E.C., Belasque Junior, J., Marcassa, L.G., “Detection of citrus canker in citrus plants using laser induced fluorescence spectroscopy.” Precision Agriculture. Vol. 10, pp. 319–330, 2009.
CrossRef
- Yang, C.M., Cheng, C.H., Chen, R.K., “Changes in spectral characteristics of rice canopy infested with brown plant hopper and leaf folder.” Crop Science. Vol. 47, pp. 329–335, 2007.
CrossRef
- Delalieux, S., van Aardt, J., Keulemans, W., Schrevens, E., Coppin, P., 2007. “Detection of biotic stress (Venturia inaequalis) in apple trees using hyperspectral data: Non-parametric statistical approaches and physiological implications.” European Journal of Agronomy. Vol. 27 pp. 130–143, 2007.
CrossRef
- Chen, B., Wang, K., Li, S., Wang, J., Bai, J., Xiao, C., Lai, J., “Spectrum characteristics of cotton canopy infected with verticillium wilt and inversion of severity level.” In IFIP International Federation for Information Processing, Volume 259; Computer and Computing Technologies in Agriculture, vol. 2, Daoliang Li, Springer, Boston, pp. 1169–1180, 2008.
CrossRef
- Choi, Y.H., Tapias, E.C., Kim, H.K., Lefeber, A.W.M., Erkelens, C., Verhoeven, J.T.J., Brzin, J., Zel, J., Verpoorte, R., “Metabolic discrimination of Catharanthus roseus leaves infected by phytoplasma using 1H-NMR spectroscopy and multivariate data analysis.” Plant Physiology. Vol. 135, pp. 2398–2410, 2004.
CrossRef
- Graeff, S., Link, J., Claupein, W., “Identification of powdery mildew (Erysiphe graminis sp. tritici) and take-all disease (Gaeumannomyces graminis sp. tritici) in wheat (Triticum aestivum L.) by means of leaf reflectance measurements.” Central European Journal of Biology. Vol. 1, pp. 275–288, 2006.
CrossRef
- Lorenzen, B., Jensen, A., “Changes in spectral properties induced in Barley by cereal Powdery Mildew.” Remote Sensing Environment. Vol. 27, pp. 201–209, 1989.
CrossRef
- Sasaki, Y., Okamoto, T., Imou, K., Torii, T., 1998. Automatic diagnosis of plant disease: spectral reflectance of healthy and diseased leaves. In: Proceedings AgEng’98 Conference, Oslo, pp. 6.
CrossRef
- Mohammad Ei –Helly, Ahmed Rafea, Salwa Ei – Gamal And Reda Abd Ei Whab., “Integrating Diagnostic Expert System With Image Processing Via Loosely Coupled Technique,” Central Laboratory for Agricultural Expert System(CLAES), 2004.
- Sungkur R., Baichoo S., “An automated system to recognize Fungi-caused diseases sugarcane leaves,” Research journal of University of Maurititus, pp.1-20, 2009.
- Shen Weizhong, Wu Yachun, “Grading method of leaf spot disease based on image processing,’’ IEEE, pp.491-494, 2008.
- Rakesh Kaundal, Amar S Kapoor and Gajendra PS Raghava, “Machine learning techniques in disease forecasting: a case study on rice blast prediction,” BMC Bioinformatics., 2006.
- Prasad Babu M. S. and Srinivasa R. B., “ Leaves Recognition Using Back Propagation Neural Network-Advice For Pest and Disease Control On Crops,” IndiaKisan.Net: Expert Advissory System, 2007.
- Alexander A. Doudkin , Alexander V. Inyutin, Albert I. Petrovsky, Maxim E. Vatkin[2007] “Three Level Neural Network for Data Clusterzation on Images of Infected Crop Field,” Journal of Research and Applications in Agricultural Engineering, Vol.52, 2007.
- Pixia, D. and Xiangdong W., “Recognition of Greenhouse Cucumber Disease Based on Image Processing Technology,” Open Journal of Applied Sciences, Vol. 3, pp. 27-31, 2013.
CrossRef
- Vijaya kumar J. V., Arumugam, S., “Recognition of Powdery Mildew Disease for Betelvine Plants Using Digital Image Processing”. International Journal of Distributed and Parallel Systems (IJDPS) Vol.3, 2012.
- Xu Pengyun & Li Jigang, “Computer assistance image processing spores counting, 2009 International Asia Conference on Informatics in Control, Automation and Robotics,” IEEE computer society, pp.203-206, 2009.
- Seiffert U. and Schweizer, P., “A Pattern Recognition Tool for Quantitative Analysis of In Planta Hyphal Growth of Powdery Mildew Fungi.” Technical Advance. Vol. 18, pp. 906-912, Sept. 2005.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.